MESC (Yao et al): How much disease heritability is mediated by gene expression?
Work led by Doug Yao on quantifying the amount of disease heritability that is mediated by gene expression QTLs is out:
Quantifying genetic effects on disease mediated by assayed gene expression levels.
D Yao, LJ O’Connor, AL Price, A Gusev. 2019
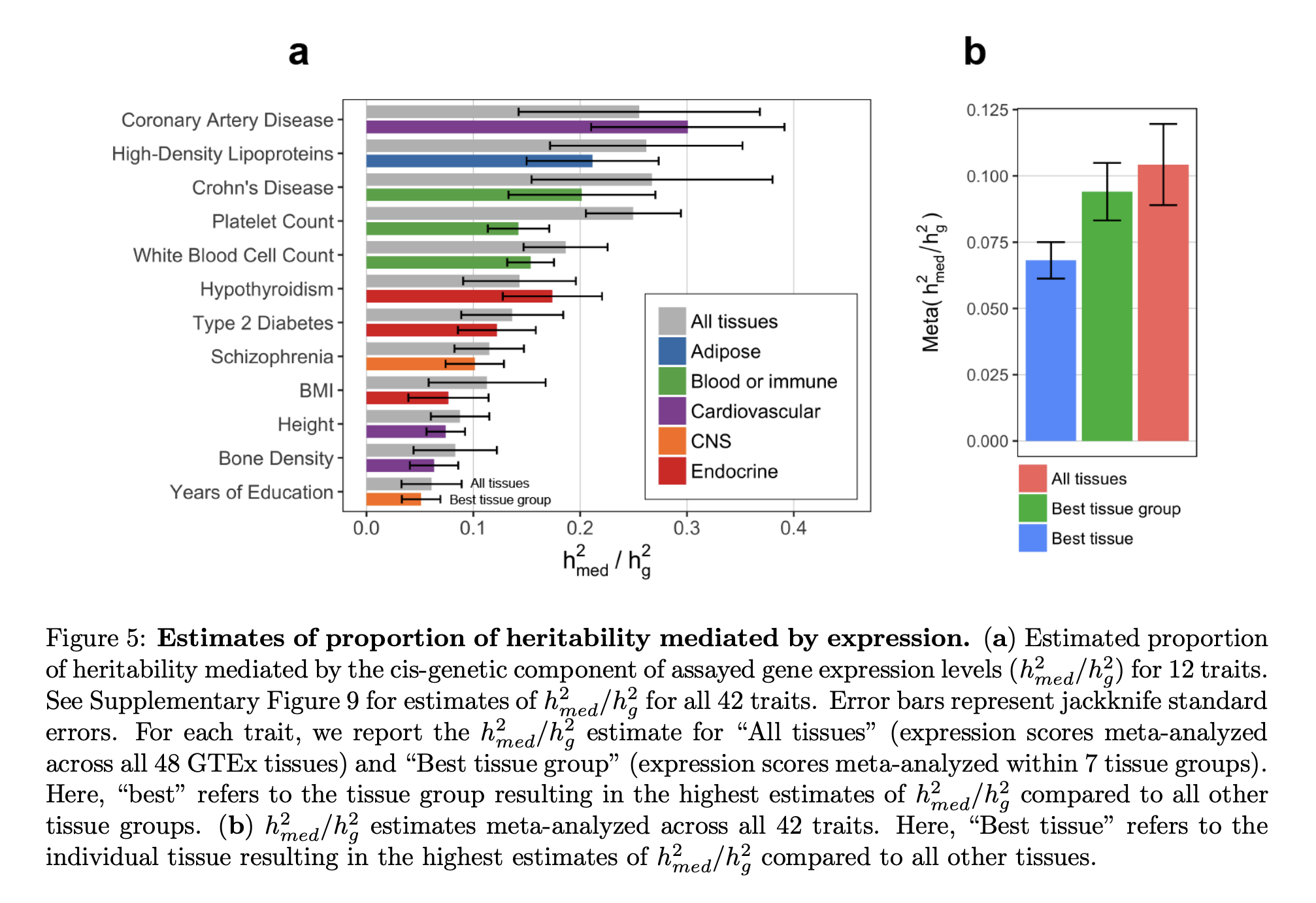
GWAS and expression integration (methods such as TWAS, mendelian randomization, colocalization, etc.) have implicated many potential mechanisms across many diseases, but it is not known whether these mechanisms are actually causally mediating the disease or just coincidentally associated with it. In this work, Douglas Yao et al. developed a novel method - Mediated Expression Score regression (MESC) - to estimate the total fraction of disease heritability that is causally mediated by all genetic effects on expression. Applying this method to gene expression from multiple tissues in the GTEx study, and dozens of complex diseases, we find that a statistically significant but low proportion of disease heritability (mean 11% s.e. 2%) is mediated by the cis-genetic component of assayed gene expression levels. We observed an inverse relationship between cis-heritability of expression and disease heritability mediated by expression, suggesting that genes with weaker eQTLs - i.e. those that are the hardest to detect - have larger causal effects on disease. We also found that expression-mediated heritability was enriched in disease relevant gene sets including loss-of-function intolerant genes and FDA-approved drug targets.
Our results demonstrate that steady-state expression levels in bulk tissues are informative of regulatory disease mechanisms, but that (on average) the majority of disease heritability remains unaccounted for by current eQTL studies. What explains this gap? Extremely weak effects on expression, trans-QTLs, cell-type or context-specificity are all possible culprits, and we investigate these possibilites in the manuscript.
The method & code is available in the MESC repository.
This work is related to recent studies of the relationship between eQTL and GWAS signal by Wang et al. and Chun et al 2017 Nat Genet, as well as the recent TWAS Perspective of Wainberg et al. 2019 Nat Genet.