PLASMA (Wang et al): A new method for allele-specific fine-mapping
Work led by Austin Wang on fine-mapping causal variants from molecular data using using allelic imbalance is now online:
Allele-Specific QTL Fine-Mapping with PLASMA.
AT Wang, AH Shetty, E O’Connor, C Bell, MM Pomerantz, ML Freedman, A Gusev. 2019
The goal of fine-mapping is to prioritize genetic variants that are likely to impact a trait, with statistical guarantees. Austin Wang developed a powerful new method – PLASMA – for fine-mapping molecular (gene expression, chromatin activity, etc.) data by using allele-specific variation observable within an individual, in addition to traditional inter-individual changes. PLASMA achieves an order of magnitude reduction in the sample size needed for accurate fine-mapping relative to existing methods. PLASMA yields higher specificity across multiple scales of functional data while enriching for features we believe to be causal. This approach formalizes the relationship between genetic variants in the population and allelic activity in the individual, with applications to other contexts such as prediction and colocalization.
The method & code is available in the PLASMA repository.
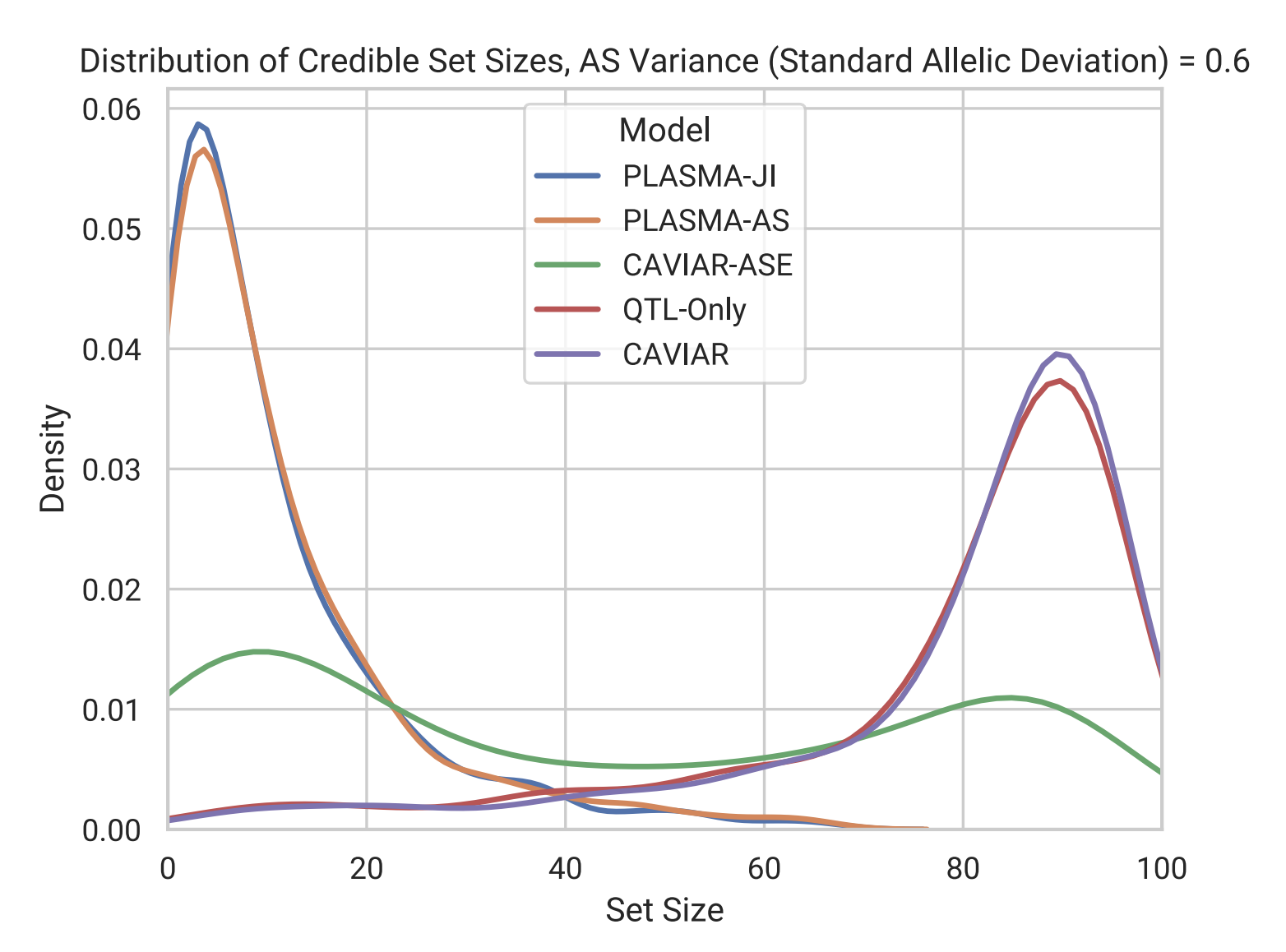
Distribution of fine-mapped SNP set sizes from various methods in simulation