A new method to infer cell-type/tumor specific QTLs (Kalita et al. pre-print)
Work led by Cynthia Kalita, developing a new method to identify cell-type specific QTLs in bulk RNA-seq data by leveraging allele-specificity is now online:
A novel method to identify cell-type specific regulatory variants and their role in cancer risk.
Kalita C, Gusev A. 2021
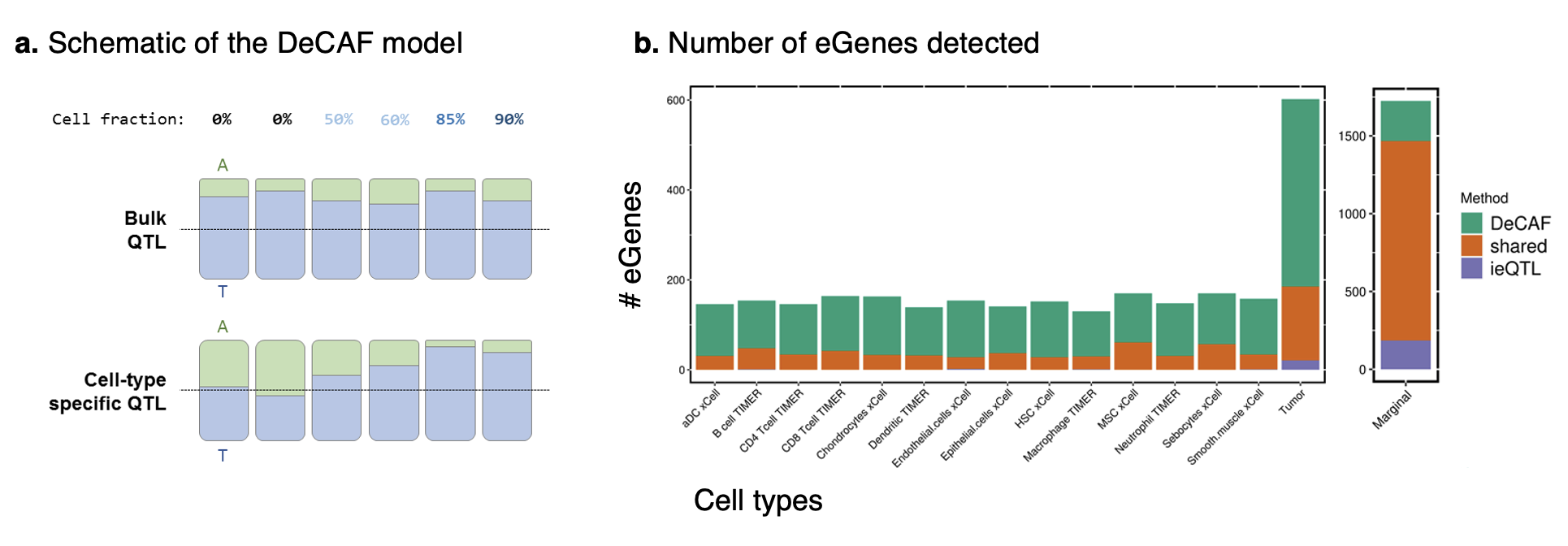
Several recent studies have shown that variants influencing gene expression in a cell-type specific manner can be detected from bulk (i.e. non-specific) RNA-seq data by leveraging differences in cell type proportions across individuals. Here, we propose the methods DeCAF to incorporate allele-specific information in addition to total expression variation, which greatly increases the power of this approach in moderately sized expression studies. DeCAF can likewise identify tumor-specific effects by leveraging differences in tumor purity. We apply DeCAF to cancer RNA-seq data from The Cancer Genome Atlas and identify a large number of specific effects that are independent of conventional eQTLs and replicate in independent datasets. Intriguingly, many of the tumor-specific effects replicate strongly in immune cell types, suggesting they may be capturing genetic variants involved in tumor-immune interactions. DeCAF can be broadly applied to cancer and non-cancer studies to identify additional interesting QTLs, and is open source.
Figure: Summary of the DeCAF model and results