Sylvan's paper on context-specific chromatin mechanisms in prostate cancer risk is out (Baca et al. Nat Genet) 🎉
Work led by Sylvan Baca on a new method to link genetic determinants of chromatin with complex disease, in collaboration with the Freedman Lab, is now out in Nature Genetics:
Genetic determinants of chromatin reveal prostate cancer risk mediated by context-dependent gene regulation.
Baca S, Singler C, Zacharia S, Seo J, Morova T, Hach F, Ding Y, Schwarz T, Huang CF, Kalita C, Groha S, Pomerantz MM, Wang V, Linder S, Sweeney CJ, Zwart W, Lack NA, Pasaniuc B, Takeda DY, Gusev A[+], Freedman ML[+]. Nature Genetics. 2022
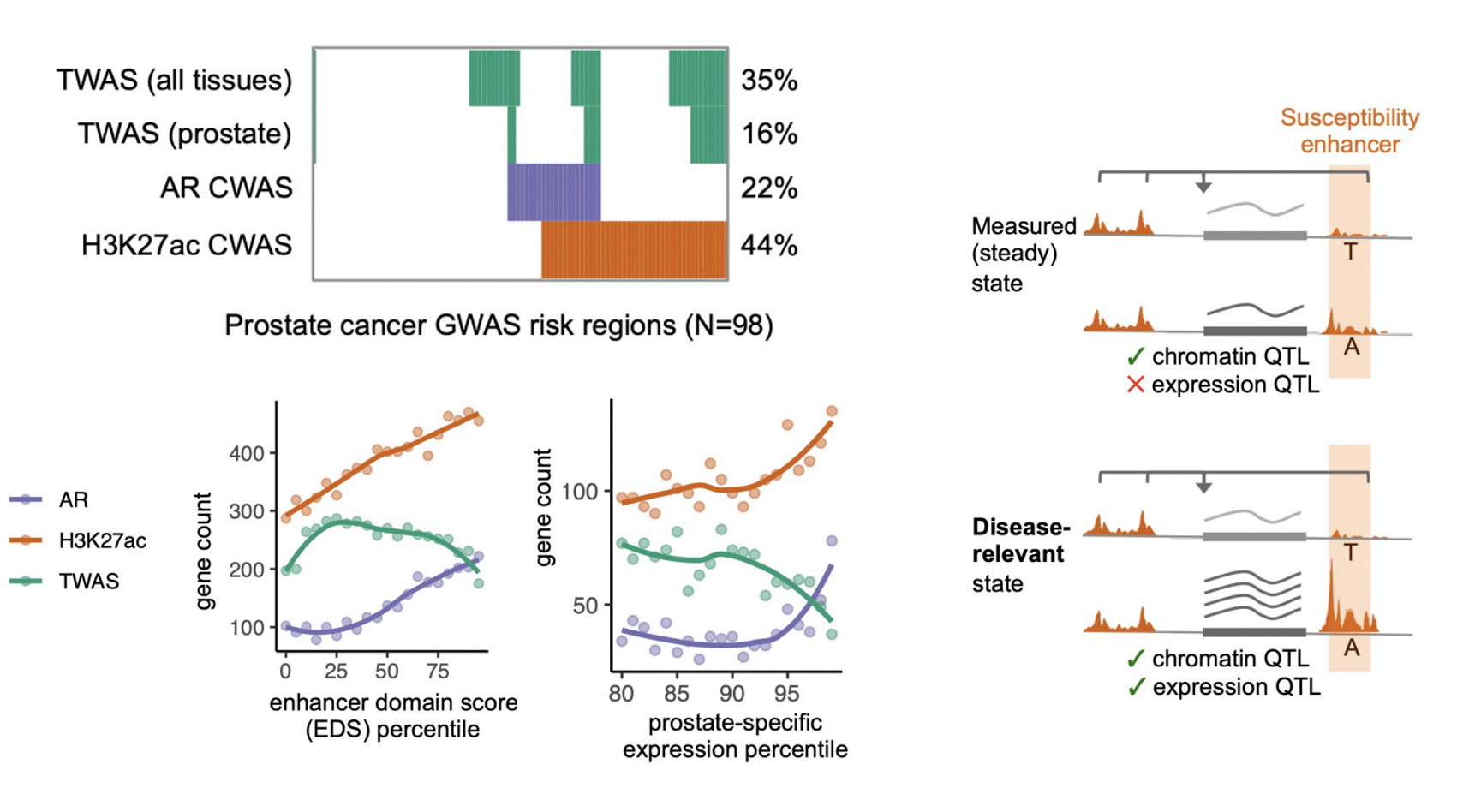
This large study of several hundred epigenomes shows that chromatin and transcription factor activity in prostate tumors is genetically controlled across thousands of regulatory elements. The genetic effect on these elements can be predicted into disease GWAS data to identify those elements associated with prostate cancer risk (which we term a “Cistrome-Wide Association Study” or CWAS). CWAS identifies many more associations than prostate transcriptomic models (echoing previous pan-cancer findings from accessibility data). Moreover, CWAS-associated elements are common at loci that lack colocalization with eQTLs, and exhibit greater cell-type specificity and evolutionary constraint; making them challenging to identify with more conventional steady-state expression data. This work adds to the growing evidence that context-specific regulatory effects may be critical to understanding complex disease.
All of the analyses in the paper could be run without directly genotyping the germline variants by instead imputing them directly from the tumor epigenomes, opening the door for CWAS analyses in many other contexts (including non-cancer).
A full analysis pipeline is available and the Baca Lab is hiring!
Figure | Summary of CWAS associations (top left); enrichment for context-dependent genes (bottom left); and hypothesized biological model (right).