Dennis' RWAS paper on epigenetic pan-cancer mechanisms is out (Grishin et al. Nat Genet) 🎉
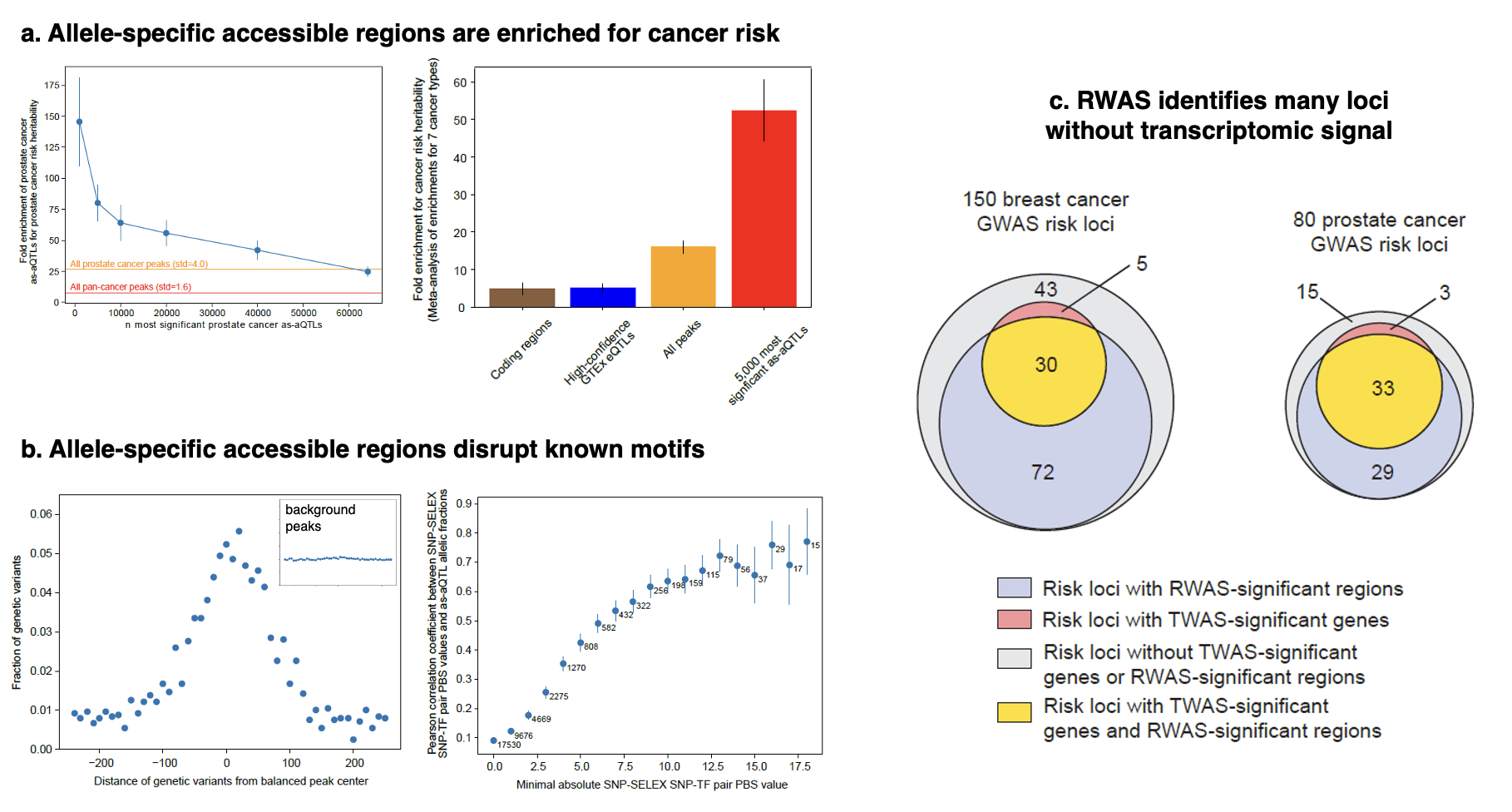
Work led by Dennis Grishin on the germline genetic mechanisms of chromatin accessibility across cancers is now out in Nature Genetics. In this work, we hypothesized that germline mutations associated with biological activity in the tumor could provide insights into these risk mechanisms; essentially working backwards from germline mutations in the cancer cell to understand the tumor initiation process. We developed a new method that combines data from hundreds of tumor epigenomes from The Cancer Genome Atlas to identify inherited mutations that influence epigenetic activity. We showed that these tumor-acting mutations extensively overlap with cancer causing mutations from Genome-Wide Association Studies (GWAS), and in fact are more significantly overlapping than any previously known class of mutations (even more so than coding variants). These variants typically disrupted oncogenic Transcription Factors and were highly concordant with effects from direct experimental perturbations.
We next developed the Regulome-Wide Association Study (RWAS), which uses genetic data to predict chromatin accessibility (the so-called “regulome”) into cancer GWAS and test for association with cancer risk. This approach implicated far more risk mechanisms than prior Transcriptome-Wide Association Studies (TWAS) from healthy individuals. For breast and prostate cancer specifically, where the most data was available, we implicated hundreds of specific regions as a resource for future experimental study. Since normal accessibility data on these samples was not available, the extent to which these mechanisms are genuinely tumor-specific remains an open question, but we believe RWAS serves as an important framework for identifying missing mechanisms for the majority of cancer risk loci.
All RWAS models are publicly available through our FUSION software, and new allele-specific models can be built with our stratAS software. And check out the write-up in the DFCI Research Blog.
Allelic imbalance of chromatin accessibility in cancer identifies candidate causal risk variants and their mechanisms.
Grishin D, Gusev A. Nature Genetics. 2022